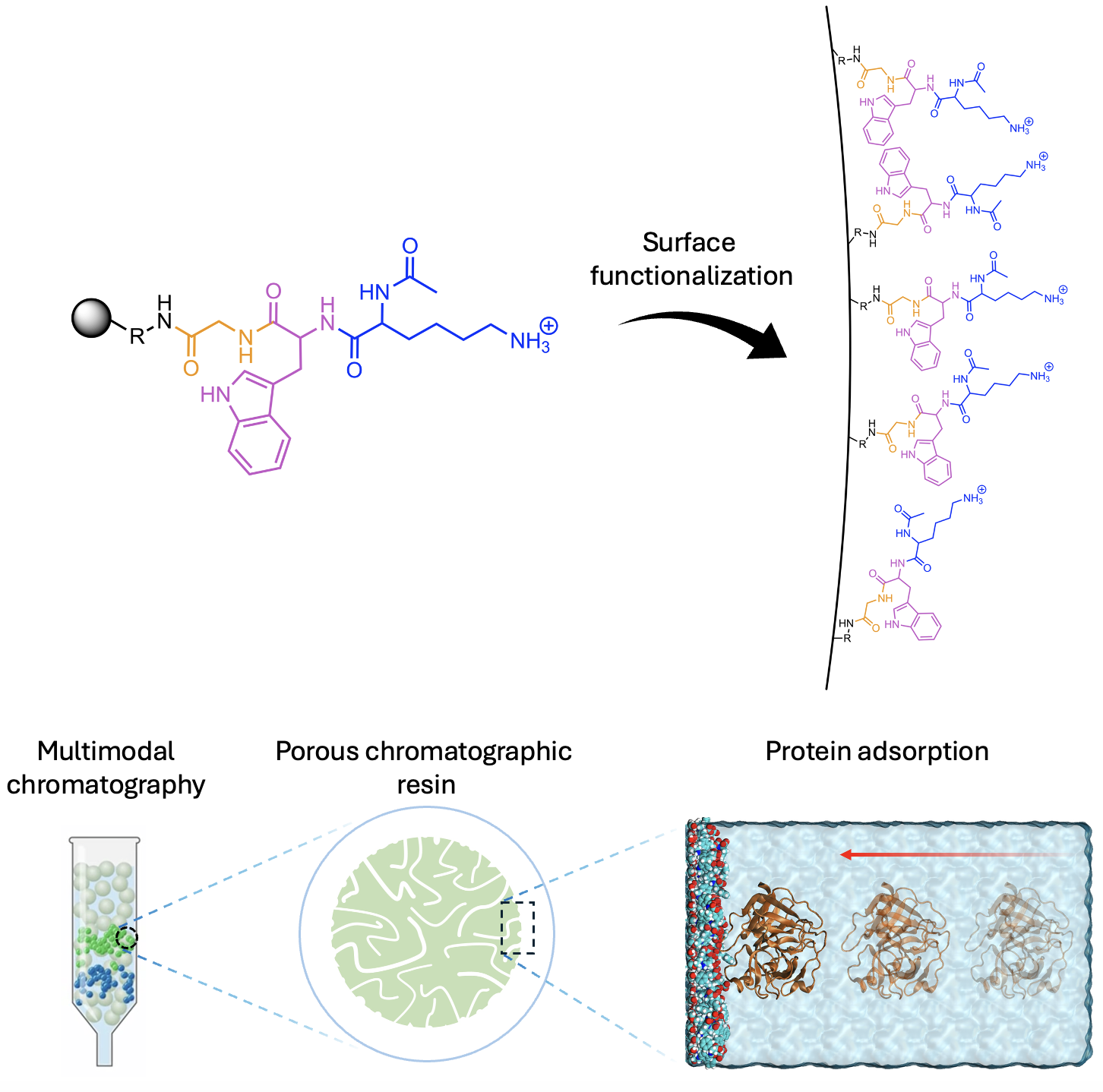
Tuning Surface Patterns and Interfacial Properties through Peptide Functionalization
Surface functionalization with peptides enables precise control of interfacial properties such as hydrophobicity and adsorption behavior in applications including chromatography, drug delivery, and tissue engineering. Noncovalent interactions between tethered peptides can create nanoscale surface patterns that significantly influence interfacial properties and are highly sensitive to subtle structural changes in the peptides, making careful peptide design essential. Through molecular dynamics simulations, we aim to understand peptide behavior on surfaces to tune interfacial properties and ultimately control adsorption, particularly in multimodal chromatography.
Applying graph neural networks to predict peptide properties
While synthetic peptides can solve problems ranging from treating antibiotic-resistant diseases to catalyzing new reactions, resource and time costs make experimentally exploring their massive design space intractable. We use a hierarchical graph approach to representing peptides in neural networks to predict properties including anti-microbial activity and chromatographic retention time.
Elucidating Biomolecular Surface Hydrophobicity
The project aims to establish design rules for peptide-functionalized surfaces, focusing on hydrophobicity as the primary objective, using Molecular Dynamics (MD) simulations and Deep Learning. These insights will aid in creating nanomaterials with tunable hydrophobicity and targeted binding affinity for specific solutes.
Injectable Pentapeptide Hydrogels for Biomimetic Applications
This project aims to design and computationally model injectable pentapeptide hydrogels that mimic the mechanical and structural properties of soft biological tissues, such as the brain. These shear-thinning, self-healing materials are developed using all-atom molecular dynamics simulations to uncover the design rules governing peptide self-assembly, hydrophobic clustering, and aromatic π-π interactions. The ultimate goal is to establish a predictive framework for designing sequence-defined materials with tunable properties. Future work will expand into coarse-grained simulations and generative modeling with diffusion-based deep learning (e.g., DDPM) to accelerate the discovery of functional peptide sequences for a wide range of biomedical applications.